Trifunctional Cross-Linker for Mapping Protein-Protein Interaction Networks and Comparing Protein Conformational States
Tan, D.; Li, Q.; Zhang, M.; Liu, C.; Ma, C.; Zhang, P.; Ding, Y.; Fan, S.; Tao, L.; Yang, B.; Li, X.; Ma, S.; Liu, J.; Feng, B.; Liu, X.; Wang, H.; He, S.; Gao, N.; Ye, K.; Dong, M.*; Lei, X.*
eLife 2016; 5:e12509
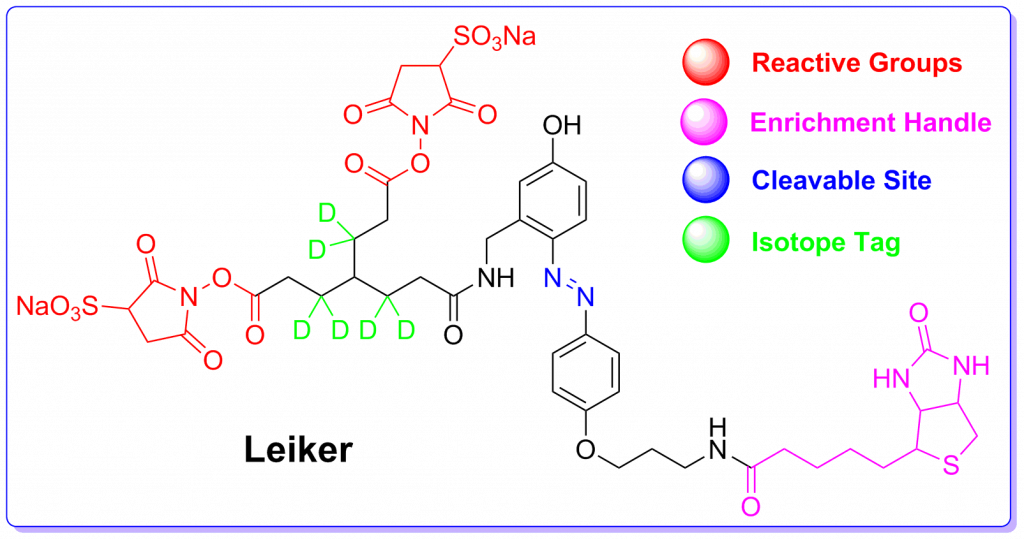
To improve chemical cross-linking of proteins coupled with mass spectrometry (CXMS), we developed a lysine-targeted enrichable cross-linker containing a biotin tag for affinity purification, a chemical cleavage site to separate cross-linked peptides away from biotin after enrichment, and a spacer arm that can be labeled with stable isotopes for quantitation. By locating the flexible proteins […]
To improve chemical cross-linking of proteins coupled with mass spectrometry (CXMS), we developed a lysine-targeted enrichable cross-linker containing a biotin tag for affinity purification, a chemical cleavage site to separate cross-linked peptides away from biotin after enrichment, and a spacer arm that can be labeled with stable isotopes for quantitation. By locating the flexible proteins on the surface of 70S ribosome, we show that this trifunctional cross-linker is effective at attaining structural information not easily attainable by crystallography and electron microscopy. From a crude Rrp46 immunoprecipitate, it helped identify two direct binding partners of Rrp46 and 15 protein-protein interactions (PPIs) among the co-immunoprecipitated exosome subunits. Applying it to?E. coli?and?C. elegans?lysates, we identified 3130 and 893 inter-linked lysine pairs, representing 677 and 121 PPIs. Using a quantitative CXMS workflow we demonstrate that it can reveal changes in the reactivity of lysine residues due to protein-nucleic acid interaction.